What's in a name? A history of taxonomy
By Sandra Knapp
Taxonomy and technology
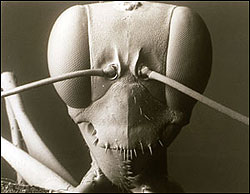
Gigantiops destructor, a tropical ant, as seen under a scanning electron microscope.
The technological advances of the twentieth century have made the process of examining organisms' characters much easier. For example, microscopes allow us to look at the shapes of chloroplasts, the bodies that convert light into energy, in particular plant cells. But one of the most significant innovations of all has been the scanning electron microscope. This microscope uses an electron beam to produce magnifying power that allows the viewer to count the bristles on an ant's nose, and to look inside the anthers of plants at the pollen, which is often fantastically sculptured and species specific in shape. Even plankton, microscopic marine organisms which were previously thought to have no clearly visible external characteristics, display a huge variety of shapes and characters under the revealing gaze of the scanning electron microscope.
Characteristics can also be studied at the cellular level. Extracts of plant or animal material containing enzymes can be placed within an electric field and different proteins will travel different distances. One can look at the banding patterns of these different proteins and analyze which populations share certain enzymes and how populations or species differ. This technique is called allozyme electrophoresis, and can be extremely useful for detecting hybridization in nature.
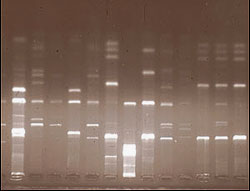
Banding patterns formed by chopped up DNA fragments from a fern.
An organism's interaction with its environment can also be helpful to the taxonomist. Behaviour has also become very important in determining taxonomic relationships and looking at how things are related to one another. Many parasitic insects are host-specific and only parasitize a single other species of plant or animal. Studying the behaviour of these interactions can give insect taxonomists clues about relationships and about the evolution of this peculiar way of life--parasitism.
The genetic code
One of the greatest technological revolutions of all has been the invention of methods enabling scientists to look directly at the genetic code. The discovery of the shape and function of DNA allows taxonomists to study the genes that code for the organism's characters.
Thinking point
Taxonomists of the past would be amazed to learn of the technologies that modern day researchers have at their disposal, and the information that those technologies reveal. Do you think that future developments will deepen our understanding of taxonomy to a level that we could not conceive of today?
All cells have DNA in the nucleus and also in their organelles, the small cellular components such as chloroplasts or mitochondria. All the DNA from plant cells for example, can be extracted, then separated into bands of different density which correspond to the different size genomes from different parts of the cell. One way of looking at DNA is to take a sequence of DNA, say the chloroplast DNA, which is much smaller than nuclear DNA, from the cells and chop it up with special enzymes. The resulting fragments can then be examined in the same way as allozymes are in an electric field. The banding patterns--the distribution of the bands--can be studied to assess the relationships between different plant species.
polymerase chain reaction (PCR), which is essentially an iterative copying procedure using bacterial enzymes, changed the practise of taxonomy completely. Using this method, a sequence of DNA is taken and, through the copying procedure, amplified to make huge amounts which can then be sequenced. The sequencing is the reading of the nucleotides that make up the DNA. The use of PCR copying means that enough DNA to sequence can be obtained from very small samples, even a square centimetre of leaf tissue can be enough, or DNA can even be obtained from old herbarium specimens!
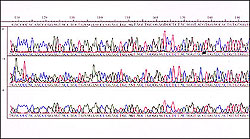
Output of a DNA sequencer
All DNA is made up of four nucleotides (deoxyribonucleotides--hence the abbreviation DNA); adenine (A) guanine (G), cytosine (C) and thymine (T). The DNA double helix consists of pairs of nucelotide bases--A always pairs with T, and G always pairs with C, which link the two strands together. Sequencers read single stranded DNA--like people read a line of text in a book. Each amino acid produced by living cells is coded for by a triplet of nucleotides; for example TTG codes for the essential amino acid leucine, while a triple with a single nucleotide difference, ATG, codes for methionine. Proteins that carry out cell functions are made up of a specific sequence of amino acids. So the sequence of the bases in the DNA has a direct link to the cellular machinery that allows all organisms to develop, grow and live in their environments.
DNA sequencing
When single stranded DNA is put through an automated sequencer, the computer produces output that looks like many overlapping lines. Each base shows up as a different colour: thymine is red, guanine is black, adenine is green and cytosine is blue. The peaks correspond to the presence of particular nucleotides and from this a sequence--ATCGTTA for example--can be read. These sequences need to be aligned to make sure they are all in the correct 'reading frame'.
Sequencing, however, is not the end of the story. To determine relationships between organisms is much more complicated than that. Patterns and sequences are sometimes extremely complex and can mislead the researcher. For example, some plants from the tobacco genus Nicotiana show two phylogenetic trees, one constructed using a sequence of DNA from the nucleus and the other a sequence from the chloroplast genome. Chloroplasts in plants are inherited maternally--so the paternal genome never shows up in a tree based on chloroplast sequences. Nicotiana contains many species that are of hybrid origin and are allopolyploid--they have twice the normal number of chromosomes, a set from one of each parent. The trees based on chloroplast and nuclear sequences conflict due to hybridity--if we did not know these species were of hybrid origin, we would be seriously confused!
Many patterns seen at the DNA level are still not understood. For example sometimes genes convert one to another or only one parental type is inherited. Nicotiana tabacum, which is cultivated tobacco, is a tetraploid hybrid species, with two sets of chromosome pairs--its two parents are different diploid species, each with one set of chromosome pairs. It inherits the chloroplast genes only from the maternal parent, Nicotiana sylvestris, and so it groups with that species on the tree based on chloroplast sequences. Tobacco groups with its paternal parent, Nicotiana tomentosiformis, on the tree based on nuclear sequences. But nuclear sequence /paternal, chloroplast sequences /maternal is not always the pattern seen. Sometimes, both sets of sequences group a hybrid species with its mother and complicated patterns can result. So sequencing does not provide all the answers as was originally hoped.

Arabidopsis thaliana from The Natural History Museum herbarium.
The genomes of some species have been sequenced completely. The only plant to have had its entire genome sequenced so far is Arabidopsis thaliana, a humble little weed in the mustard family with very simple flowers. The taxonomy of Arabidopsis is not yet well understood, despite us knowing all about this one species. To really determine relationships and study evolutionary patterns in nature, we must know about more than one thing. Ideally one would need three characters, to construct a three taxon hypothesis as described earlier.
Botanical taxonomists have long been interested in the development of flowers because many sexual characters, which are found in the flowers, are used to classify plants. Researchers are now looking at the genes that control flower development, at the ways they are turned on and off and how they relate to each other. What is being sought is essentially the genetic basis for what Linnaeus called the sexual system over 300 years ago.
Toolbox

Our scientists study the snails that host the schistosomiasis parasite, which causes a disease that affects nearly 200 million people.
